Abstract
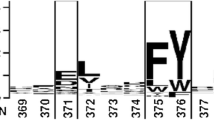
Appropriate amino acid substitutions are critical for protein engineering to redesign catalytic properties of industrially important enzymes like lipases. The present study aimed for improving the environmental stability of lipase from Pseudomonas plecoglossicida S7 through site-directed mutagenesis driven by computational studies. lipA gene was amplified and sequenced. Both wild type (WT) and mutant type (MT) lipase genes were expressed into the pET SUMO system. The expressed proteins were purified and characterized for pH and thermostability. The lipase gene belonged to subfamily I.1 lipase. Molecular dynamics revealed that Y12F-palmitic acid complex had a greater binding affinity (-6.3 Kcal/mol) than WT (-6.0 Kcal/mol) complex. Interestingly, MDS showed that the binding affinity of WT-complex (-130.314 ± 15.11 KJ/mol) was more than mutant complex (-108.405 ± 69.376 KJ/mol) with a marked increase in the electrostatic energy of mutant (-26.969 ± 12.646 KJ/mol) as compared to WT (-15.082 ± 13.802 KJ/mol). Y12F mutant yielded 1.27 folds increase in lipase activity at 55 °C as compared to the purified WT protein. Also, Y12F mutant showed increased activity (~ 1.2 folds each) at both pH 6 and 10. P. plecoglossicida S7. Y12F mutation altered the kinetic parameters of MT (Km- 1.38 mM, Vmax- 22.32 µM/min) as compared to WT (Km- 1.52 mM, Vmax- 29.76 µM/min) thus increasing the binding affinity of mutant lipase. Y12F mutant lipase with better pH and thermal stability can be used in biocatalysis.







Similar content being viewed by others
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Abraham MJ, Murtola T, Schulz R et al (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25. https://doi.org/10.1016/j.softx.2015.06.001
Akbari N, Khajeh K, Rezaie S et al (2010) High-level expression of lipase in Escherichia coli and recovery of active recombinant enzyme through in vitro refolding. Protein Expr Purif 70(1):75–80. https://doi.org/10.1016/j.pep.2009.08.009
Akbulut N, Öztürk MT, Pijning T, Öztürk S?, Gümüşel F (2013) Improved activity and thermostability of Bacillus pumilus lipase by directed evolution. Journal of biotechnology. 10;164(1):123-9. https://doi.org/10.1016/j.jbiotec.2012.12.016
Akdel M, Pires DEV, Pardo EP et al (2021) A structural biology community assessment of AlphaFold 2 applications. BioRxiv, https://doi.org/10.1101/2021.09.26.461876
Ali Y, Ahmad B, Alghamdi KM, Kamal T, Ali HS, Anwar Y, Hussain A, Jogezai NU et al (2019) Characterization of recombinant cold active lipase from a novel Pseudomonas spp. MG687270.International Journal of Agriculture and Biology.22(5):855–65. https://doi.org/10.17957/IJAB/15.1141
Anderson RJ, Weng Z, Campbell RK, Jiang X (2005) Main-chain conformational tendencies of amino acids. Proteins: Struct Funct Genet 60(4):679–689. https://doi.org/10.1002/prot.20530
Arber W (2000) Genetic variation: molecular mechanisms and impact on microbial evolution. FEMS Microbiol Rev 24(1):1–7. https://doi.org/10.1111/j.1574-6976.2000.tb00529.x
Beselin A (2005) Optimization of lipase production in Burkholderia glumae. Patent-Nr, vol 56817. BASF AG (Ludwigshafen)
Bhatwa A, Wang W, Hassan YI et al (2021) Challenges associated with the formation of recombinant protein inclusion bodies in Escherichia coli and strategies to address them for industrial applications. Front Bioeng Biotechnol 10:65. https://doi.org/10.3389/fbioe.2021.630551
Börjesson U, Hünenberger PH (2003) Effect of mutations involving charged residues on the stability of Staphylococcal nuclease: a continuum electrostatics study. Protein Eng Des Sel 16(11):831–840. https://doi.org/10.1093/protein/gzg103
Braberg H, Echeverria I, Kaake RM et al (2022) From systems to structure—using genetic data to model protein structures. Nat Rev Genet 23:342–354. https://doi.org/10.1038/s41576-021-00441-w
Chandra P, Enespa, Singh R et al (2020) Microbial lipases and their industrial applications: a comprehensive review. Microb Cell Factories 19:169. https://doi.org/10.1186/s12934-020-01428-8
Choubey A, Dehury B, Kumar S et al (2020) Naltrexone a potential therapeutic candidate for COVID-19. J Biomol Struct Dyn 40(3):963–970. https://doi.org/10.1080/07391102.2020.1820379
Chen H, Yu F, Shi N et al (2021) Overexpression and Mutation of a Novel Lipase from Serratia marcescens L1 in Escherichia coli. Process Biochemistry. 111:233 – 40. https://doi.org/10.1016/j.procbio.2021.11.001
Choudhary P, Bhowmik A, Verma S et al (2022a) Multi-substrate sequential optimization, characterization and immobilization of lipase produced by Pseudomonas plecoglossicida S7. Environ Sci Pollut Res 16:1–5.https://doi.org/10.1007/s11356-022-22098-6
Choudhary P, Bhowmik A, Chakdar H et al (2022b) Understanding the biological role of PqqB in Pseudomonas stutzeri using molecular dynamics simulation approach. J Biomol Struct Dyn 40(9):4237–4249.https://doi.org/10.1080/07391102.2020.1854860
Contesini FJ, Davanço MG, Borin GP et al (2020) Advances in recombinant lipases: production, engineering, immobilization and application in the pharmaceutical industry. Catalysts 10(9):1032. https://doi.org/10.3390/catal10091032
DeLano WL (2014) The PyMOL Molecular Graphics System, Version 1.8. Schrödinger LLC
Dobson CM (2019) Biophysical techniques in structural biology. Annu Rev Biochem 88:25–33. https://doi.org/10.1146/annurev-biochem-013118-111947
Dürauer A, Mayer S, Sprinzl W et al (2009) High-throughput system for determining dissolution kinetics of inclusion bodies. Biotechnol J 4(5):722–729. https://doi.org/10.1002/biot.200800290
Fazil MH, Kumar S, Farmer R et al (2011) Binding efficiencies of carbohydrate ligands with different genotypes of cholera toxin B: molecular modeling, dynamics and Docking Simulation studies. J Mol Model 18(1):1–10. doi: https://doi.org/10.1007/s00894-010-0947-6
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evol (New York) 39(4):783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Fernández-Lorente G, Palomo JM, Fuentes M et al (2003) Self‐assembly of Pseudomonas fluorescens lipase into bimolecular aggregates dramatically affects functional properties. Biotechnol Bioeng 82(2):232–237. https://doi.org/10.1002/bit.10560
Fields PA, Dong Y, Meng X et al (2015) Adaptations of protein structure and function to temperature: there is more than one way to ‘skin a cat’. J Exp Biol 218(12):1801–1811. https://doi.org/10.1242/jeb.114298
Futagi Y, Kobayashi M, Narumi K et al (2019) Homology modeling and site-directed mutagenesis identify amino acid residues underlying the substrate selection mechanism of human monocarboxylate transporters 1 (hMCT1) and 4 (hMCT4). Cell Mol Life Sci 76:4905–4921. https://doi.org/10.1007/s00018-019-03151-z
Girdhar K, Dehury B, Singh MK et al (2018) Novel insights into the dynamics behavior of glucagon-like peptide-1 receptor with its small molecule agonists. J Biomol Struct Dyn 37(15):3976–3986. https://doi.org/10.1080/07391102.2018.1532818
Glynou K, Ioannou PC, Christopoulos TK (2003) One-step purification and refolding of recombinant photoprotein aequorin by immobilized metal-ion affinity chromatography. Protein Expr Purif 27(2):384–390. https://doi.org/10.1016/S1046-5928(02)00614-9
Guan L, Gao Y, Li J et al (2020) Directed evolution of Pseudomonas fluorescens lipase variants with improved thermostability using error-prone PCR. Front Bioeng Biotechnol 8:1034. https://doi.org/10.3389/fbioe.2020.602138
Gupta N, Rathi P, Gupta R (2002) Simplified para-nitrophenyl palmitate assay for lipases and esterases. Anal Biochem 311(1):98–99. https://doi.org/10.1016/S0003-2697(02)00379-2
Hamdan SH, Maiangwa J, Ali MS et al (2021) Thermostable lipases and their dynamics of improved enzymatic properties. Appl Microbiol Biotechnol 105(19):7069–7094. https://doi.org/10.1007/s00253-021-11520-7
Heater BS, Chan WS, Lee MM, Chan MK (2019) Directed evolution of a genetically encoded immobilized lipase for the efficient production of biodiesel from waste cooking oil. Biotechnol Biofuels 12:165. https://doi.org/10.1186/s13068-019-1509-5
Johnson AC, Reitz M, Greene JJ, Venigalla B, Rao (2021) CRC press,Boca Raton. 21. https://doi.org/10.1201/9781003067658
Jumper J, Evans R, Pritzel A et al (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. https://doi.org/10.1038/s41586-021-03819-2
Karakaş F, Arslanoğlu A (2020) Gene cloning, heterologous expression, and partial characterization of a novel cold-adapted subfamily I. 3 lipase from Pseudomonas fluorescence KE38. Sci Rep 10:22063. https://doi.org/10.1038/s41598-020-79199-w
Kaur J, Kumar A, Kaur J (2018) Strategies for optimization of heterologous protein expression in E. coli: Roadblocks and reinforcements. Int. J. Biol. Macromol., 106:803–822. https://doi.org/10.1016/j.ijbiomac.2017.08.080Kawaguchi M, Yasumasu S, Shimizu A,. (2013) Adaptive evolution of fish hatching enzyme: one amino acid substitution results in differential salt dependency of the enzyme. J. Exp. Biol., 216 (9):1609–1615.https://doi.org/10.1242/jeb.069716
Kawase M, Takahashi H, Sonomoto K et al (1991) Effect of chemical modification of tyrosine residues on activities of bacterial lipase. J Ferment Bioeng 72(5):317–319. https://doi.org/10.1016/0922-338X(91)90079-V
Kibbe WA (2007) OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res 35:W43–W46 (Issue Supplement 2. doi:https://doi.org/10.1093/nar/gkm234
Kovacic F, Babic N, Krauss U et al (2019) Classification of lipolytic enzymes from bacteria. Handbook of hydrocarbon and lipid Microbiology. Editor: Fernando Rojo, vol 24. Springer Cham, Switzerland, pp 255–289. https://doi.org/10.1007/978-3-319-50418-6
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1974. https://doi.org/10.1093/molbev/msw054
Kumari R, Kumar R, Consortium OSDD et al (2014) g_mmpbsa- A GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54(7):1951–1962. https://doi.org/10.1021/ci500020m
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227(5259):680–685. https://doi.org/10.1038/227680a0
Law MJ, Linde ME, Chambers EJ et al (2006) The role of positively charged amino acids and electrostatic interactions in the complex of U1A protein and U1 hairpin II RNA. Nucleic Acids Res 34(1):275–285. https://doi.org/10.1093/nar/gkj436
Hans L, Burk D (1934) The determination of enzyme dissociation 559 constants. J Am Chem Soc 563:658–666
Li C, Zhang R, Wang J et al (2020) Protein engineering for improving and diversifying natural product biosynthesis. Trends in biotechnology 38(7):729–744. https://doi.org/10.1016/j.tibtech.2019.12.008
Liao M, Somero GN, Dong Y (2019) Comparing mutagenesis and simulations as tools for identifying functionally important sequence changes for protein thermal adaptation. Proc. Natl. Acad. Sci. U.S.A., 116 (2):679–688. https://doi.org/10.1073/pnas.181745511
Luo C, Hu X, Bao M et al (2022) Efficient biodegradation of phenanthrene using Pseudomonas stutzeri LSH-PAH1 with the addition of sophorolipids: alleviation of biotoxicity and co-metabolism studies. Environ Pollut 301:119011. https://doi.org/10.1016/j.envpol.2022.119011
Lüthy R, Bowie JU, Eisenberg D (1992) Assessment of protein models with three-dimensional profiles. Nature 356:83–85.https://doi.org/10.1038/356083a0
Madan B, Mishra P (2010) Co-expression of the lipase and foldase of Pseudomonas aeruginosa to a functional lipase in Escherichia coli. Appl Microbiol Biotechnol 85:597–604. https://doi.org/10.1007/s00253-009-2131-4
Mohammadi M, Sepehrizadeh Z, Ebrahim-Habibi A et al (2016) Enhancing activity and thermostability of lipase A from Serratia marcescens by site-directed mutagenesis. Enzyme Microb Technol 93–94:18–28. https://doi.org/10.1016/j.enzmictec.2016.07.006
Nantel G, Proulx P (1973) Lipase activity in E. coli. Biochim Biophys 316(2):156–161. https://doi.org/10.1016/0005-2760(73)90005-2
Patra AK, Mukhopadhyay R, Mukhija R et al (2000) Optimization of inclusion body solubilization and renaturation of recombinant human growth hormone from Escherichia coli. Protein Expre Purif 18(2):182–192. https://doi.org/10.1006/prep.1999.1179
Pettersen EF, Goddard TD, Huang CC et al (2004) UCSF Chimera - A visualization system for exploratory research and analysis. J Comput Chem 25(13):1605–1612. https://doi.org/10.1002/jcc.20084
Phukon LC, Chourasia R, Kumari M et al (2020) Production and characterisation of lipase for application in detergent industry from a novel Pseudomonas helmanticensis HS6. Bioresour. Technol., 309: 123352. https://doi.org/10.1016/j.biortech.2020.123352
Priyanka P, Kinsella G, Henehan GT et al (2019) Isolation, purification and characterization of a novel solvent stable lipase from Pseudomonas reinekei. Protein Expre Purif 153:121–130. https://doi.org/10.1016/j.pep.2018.08.007
Raftari M, Ghafourian S, Bakar FA (2013) Metabolic engineering of Lactococcus lactis influence of the overproduction of lipase enzyme. J Dairy Res 80(8):490–495. https://doi.org/10.1017/S0022029913000435
Ramón A, Señorale M, Marín M (2014) Inclusion bodies: not that bad… Front. Microbiol 5:56. https://doi.org/10.3389/fmicb.2014.00056
Rinas U, Garcia-Fruitós E, Corchero JL et al (2017) Bacterial inclusion bodies: discovering their better half. Trends Biochem Sci 42(9):726–737. https://doi.org/10.1016/j.tibs.2017.01.005
Rios NS, Pinheiro BB, Pinheiro MP et al (2018a) Biotechnological potential of lipases from Pseudomonas: sources, properties and applications. Process Biochem 75:99–120.https://doi.org/10.1016/j.procbio.2018a.09.003
Rios NS, Pinheiro BB, Pinheiro MP et al (2018b) Biotechnological potential of lipases from Pseudomonas: sources, properties and applications. Process Biochem 75:99–120.https://doi.org/10.1016/j.procbio.2018b.09.003
Rosenau F, Jaeger K-E (2000) Bacterial lipases from Pseudomonas: regulation of gene expression and mechanisms of secretion. Biochimie 82(11):1023–1032. https://doi.org/10.1016/S0300-9084(00)01182-2
Rozi MFAM, Rahman RNZRA, Leow ATC et al (2021) Ancestral sequence Reconstruction of Ancient lipase from Family I. 3 bacterial lipolytic enzymes. Mol Phylogenet Evol 168:107381. https://doi.org/10.1016/j.ympev.2021.107381
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Sankar S, Ponnuraj K (2020) Less explored plant lipases: modeling and molecular dynamics simulations of plant lipases in different solvents and temperatures to understand structure-function relationship. Int J Biol Macromol 164:3546–3558. https://doi.org/10.1016/j.ijbiomac.2020.08.227
Sawano H, Koumoto Y, Ohta K et al (1992) Efficient in vitro folding of the three-disulfide derivatives of hen lysozyme in the presence of glycerol. FEBS Lett 303(1):11–14. https://doi.org/10.1016/0014-5793(92)80466-T
Silverman AD, Karim AS, Jewett MC (2020) Cell-free gene expression: an expanded repertoire of applications. Nat Rev Genet 21:151–170. https://doi.org/10.1038/s41576-019-0186-3
Singh RK, Tiwari MK, Singh R, Lee JK (2013) From protein engineering to immobilization: promising strategies for the upgrade of industrial enzymes. Int J Mol Sci 14(1):1232–1277. https://doi.org/10.3390/ijms14011232
Singh A, Upadhyay V, Upadhyay AK et al (2015) Protein recovery from inclusion bodies of Escherichia coli using mild solubilization process. Microb Cell Factories 14:41. https://doi.org/10.1186/s12934-015-0222-8
Somero GN, Lockwood BL, Tomanek L (2017) Biochemical adaptation: response to environmental challenges, from life’s origins to the Anthropocene. Sinauer Associates, Incorporated Publishers
Stank A, Kokh DB, Fuller JC et al (2016) Protein binding pocket dynamics. Acc Chem Res 49(5):809–815. https://doi.org/10.1021/acs.accounts.5b00516
Tan MS, Teh YH, Ho KL et al (2020) An application of pET SUMO protein expression system in Escherichia coli: Cloning, expression, purification, and Characterisation of native Kras4BG12V Oncoprotein. Protein J 39:54–61. https://doi.org/10.1007/s10930-019-09872-1
Tian M, Fu J, Wang Z et al (2021) Enhanced activity and stability of Rhizomucor miehei lipase by mutating N-linked glycosylation site and its application in biodiesel production. Fuel 15:304:121514. https://doi.org/10.1016/j.fuel.2021.121514
Tielen P, Kuhn H, Rosenau F et al (2013) Interaction between extracellular lipase LipA and the polysaccharide alginate of Pseudomonas aeruginosa. BMC Microbiol 13:159. https://doi.org/10.1186/1471-2180-13-159
Tripathi NK, Shrivastava A (2019) Recent developments in bioprocessing of recombinant proteins: expression hosts and process development. Front. Bioeng. Biotechnol. 2019; 7:420. https://doi.org/10.3389/fbioe.2019.00420
Vallejo LF, Brokelmann M, Marten S et al (2002) Renaturation and purification of bone morphogenetic protein-2 produced as inclusion bodies in high-cell-density cultures of recombinant Escherichia coli. J Biotechnol 94(2):185–194. https://doi.org/10.1016/S0168-1656(01)00425-4
Wang S, Xu Y, Yu X-W (2021) A phenylalanine dynamic switch controls the interfacial activation of Rhizopus chinensis lipase. Int J Biol Macromol 173:1–12. https://doi.org/10.1016/j.ijbiomac.2021.01.086
Webb B, Sali A (2016) Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics. https://doi.org/10.1002/cpbi.3
Webb B, Sali A (2017) Protein structure modeling with MODELLER. In: Kaufmann, M., Klinger, C., Savelsbergh, A. (eds) Functional Genomics. Methods in Molecular Biology, 2017; 1654. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-7231-9_4
Wicka-Grochocka M, Cieśliński H, Wanarska M (2021) Cloning, expression in Komagataella phaffii, and biochemical characterization of recombinant sequence variants of Pseudomonas sp. S9 GDSL-esterase. Acta Biochim Pol 68(3):411–417. https://doi.org/10.18388/abp.2020_5730
YuN WJ, Laird M, Melli G et al (2017) PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26(13):1608–1615. https://doi.org/10.1093/bioinformatics/btq249
Zhang A, Gao R, Diao N et al (2009) Cloning, expression and characterization of an organic solvent tolerant lipase from Pseudomonas fluorescens JCM5963. J Mol Catal 56(2–3):78–84. https://doi.org/10.1016/j.molcatb.2008.06.021
Zhang Z, Zhang X (2021) Evolution of Subfamily I. 1 Lipases in Pseudomonas aeruginosa. Curr Microbiol 78:3494–3504. https://doi.org/10.1007/s00284-021-02589-4
Zhang H, Liu H, Zhang Y et al (2019) Engineered variants of a lipase from Yarrowia lipolytica with improved trypsin resistance for enzyme replacement therapy. Protein Eng Des Sel 32(8):375–383. https://doi.org/10.1093/protein/gzaa001
Zhao J, Frauenkron-Machedjou VJ, Fulton A et al (2018) Unraveling the effects of amino acid substitutions enhancing lipase resistance to an ionic liquid: a molecular dynamics study. Phys Chem Chem Phys 20(18):9600–9609. https://doi.org/10.1039/C7CP08470F
Zhao J, Ma M, Zeng Z et al (2021) Production, purification and biochemical characterisation of a novel lipase from a newly identified lipolytic bacterium Staphylococcus caprae NCU S6. J Enzyme Inhib Med Chem 36(1):249–257. https://doi.org/10.1080/14756366.2020.1861607
Zhou H-X, Pang X (2018) Electrostatic interactions in protein structure, folding, binding, and condensation. Chem Rev 118(4):1691–1741. https://doi.org/10.1021/acs.chemrev.7b00305
Acknowledgements
The authors are grateful to ICAR-NBAIM, Mau for financial assistance and infrastructural facilities provided during the work. This is a part of Ph.D. work of the first author under supervision of the corresponding author as co-supervisor.
Funding
Authors duly acknowledge the financial support from Indian Council of Agricultural Research (ICAR).
Author information
Authors and Affiliations
Contributions
HC conceptualized the study, edited and revised the manuscript. PC executed all experimental part along with protein modeling, docking studies, and prepared the first draft of the manuscript. MW, SK and NS performed the Molecular dynamic simulation experiments and analysis. HC, SK, and SS finalized the manuscript in its final form.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
All the authors have read the manuscript and approved its submission. Authors declare no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Choudhary, P., Waseem, M., Kumar, S. et al. Y12F mutation in Pseudomonas plecoglossicida S7 lipase enhances its thermal and pH stability for industrial applications: a combination of in silico and in vitro study. World J Microbiol Biotechnol 39, 75 (2023). https://doi.org/10.1007/s11274-023-03518-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-023-03518-2